Step 1: Axial Compression#
This example involves a static simulation that solves for the deformation from axial compression. Fig. 100 shows the boundary conditions on the domain.
Fig. 100 Boundary conditions for axial compression in the x direction with roller boundary conditions on the -y, +y, and -z boundaries.#
Features
Tetrahedral cells
pylith.meshio.MeshIOCubit
pylith.problems.TimeDependent
pylith.meshio.OutputSolnBoundary
pylith.meshio.DataWriterHDF5
pylith.bc.DirichletTimeDependent
pylith.bc.ZeroDB
spatialdata.geocoords.CSGeo
pylith.materials.Elasticity
pylith.materials.IsotropicLinearElasticity
spatialdata.spatialdb.SimpleDB
Static simulation
Simulation parameters#
The parameters specific to this example are in step01_axialdisp.cfg and include:
pylithapp.metadataMetadata for this simulation. Even when the author and version are the same for all simulations in a directory, we prefer to keep that metadata in each simulation file as a reminder to keep it up-to-date for each simulation.pylithappParameters defining where to write the output.pylithapp.problemParameters for the solution field with displacement and Lagrange multiplier subfields.
We override the parameters for the Dirichlet displacement boundary conditions on the -x and +x boundaries.
We replace the ZeroDB spatial database for zero displacement values with a UniformDB to impose axial compression with 2.0 m of displacement on the two boundaries.
For a more efficient solve we use the PETSc default solver options for elasticity in parallel;
for larger simulations these are sometimes more efficient than the defaults for running in serial.
$ pylith step01_axialdisp.cfg mat_elastic.cfg
# The output should look something like the following.
>> /software/py38-venv/pylith-opt/lib/python3.8/site-packages/pylith/meshio/MeshIOObj.py:44:read
-- meshiocubit(info)
-- Reading finite-element mesh
>> /src/cig/pylith/libsrc/pylith/meshio/MeshIOCubit.cc:157:void pylith::meshio::MeshIOCubit::_readVertices(pylith::meshio::ExodusII&, pylith::scalar_array*, int*, int*) const
-- meshiocubit(info)
-- Component 'reader': Reading 24824 vertices.
>> /src/cig/pylith/libsrc/pylith/meshio/MeshIOCubit.cc:217:void pylith::meshio::MeshIOCubit::_readCells(pylith::meshio::ExodusII&, pylith::int_array*, pylith::int_array*, int*, int*) const
-- meshiocubit(info)
-- Component 'reader': Reading 134381 cells in 4 blocks.
# -- many lines omitted --
>> /src/cig/pylith/libsrc/pylith/meshio/MeshIO.cc:94:void pylith::meshio::MeshIO::read(pylith::topology::Mesh*)
-- meshiocubit(info)
-- Component 'reader': Domain bounding box:
(-460000, 340000)
(-400000, 400000)
(-400000, 2.91038e-11)
# -- many lines omitted --
>> /src/cig/pylith/libsrc/pylith/utils/PetscOptions.cc:235:static void pylith::utils::_PetscOptions::write(pythia::journal::info_t&, const char*, const pylith::utils::PetscOptions&)
-- petscoptions(info)
-- Setting PETSc options:
ksp_atol = 1.0e-12
ksp_converged_reason = true
ksp_error_if_not_converged = true
ksp_rtol = 1.0e-12
pc_type = gamg
snes_atol = 1.0e-9
snes_converged_reason = true
snes_error_if_not_converged = true
snes_monitor = true
snes_rtol = 1.0e-12
ts_error_if_step_fails = true
ts_monitor = true
ts_type = beuler
>> /software/py38-venv/pylith-opt/lib/python3.8/site-packages/pylith/problems/TimeDependent.py:139:run
-- timedependent(info)
-- Solving problem.
0 TS dt 0.01 time 0.
0 SNES Function norm 1.122453626786e+01
Linear solve converged due to CONVERGED_ATOL iterations 17
1 SNES Function norm 6.926560000115e-11
Nonlinear solve converged due to CONVERGED_FNORM_ABS iterations 1
1 TS dt 0.01 time 0.01
>> /software/py38-venv/pylith-opt/lib/python3.8/site-packages/pylith/problems/Problem.py:201:finalize
-- timedependent(info)
-- Finalizing problem.
At the beginning of the output written to the terminal, we see that PyLith is reading the mesh using the MeshIOCubit reader and that it found the domain to extend from -460 km to +340 km in the x direction, from -400 km to +400 km in the y direction, and from -400 km to 0 in the z direction.
The output also includes the scales used for nondimensionalization and the default PETSc options.
At the end of the output written to the terminal, we see that the solver advanced the solution 1 time step (static simulation).
The linear solve converged after 85 iterations and the norm of the residual met the absolute convergence tolerance (ksp_atol) .
The nonlinear solve converged in 1 iteration, which we expect because this is a linear problem, and the residual met the absolute convergence tolerance (snes_atol).
Visualizing the results#
The output directory contains the simulation output.
Each “observer” writes its own set of files, so the solution over the domain is in one set of files, the boundary condition information is in another set of files, and the material information is in yet another set of files.
The HDF5 (.h5) files contain the mesh geometry and topology information along with the solution fields.
The Xdmf (.xmf) files contain metadata that allow visualization tools like ParaView to know where to find the information in the HDF5 files.
To visualize the data using ParaView or Visit, load the Xdmf files.
In Fig. 101 we use ParaView to visualize the x displacement field using the viz/plot_dispwarp.py Python script.
We start ParaView from the examples/subduction-3d directory and then run the viz/plot_dispwarp.py Python script as described in ParaView Python Scripts.
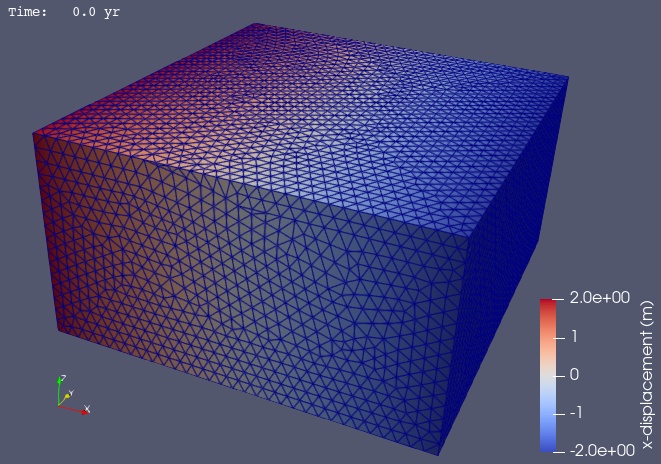
Fig. 101 Solution for Step 1. The colors of the shaded surface indicate the magnitude of the x displacement, and the deformation is exaggerated by a factor of 1000.#
